Research
Research is most fruitful (and most fun) at the intersection of different fields of research. At the core of our research is the development of methods that provide explanations for complex machine learning models, as well as the development of inherently interpretable machine learning models. We are particularly interested in getting a better understanding of how information is encoded and propagated inside a neural network, nowadays coined mechanistic interpretability, or to inform neuro-symbolic architecture designs.
Our research spans from foundations to applications in practice. One focus lies on extracting new scientific insights from models that often surpass human experts through methods for Interpretability of Machine Learning models. In molecular biology we for example use explanations of models to get new knowledge about the complex genomic mechanisms driving cancer or aging. In Neuroscience, we are interested in studying the brain through understanding a artificial neural model encoding brain activations, for example to locate and understand the processing of visual stimuli in the brain.
We further study the Explainability of models in challenging biomedical tasks, such as biomedical imaging in cancer, where an understanding of the decision-making process of a model is essential for a model to be deployed and trusted in practice.

Global Explanations
Post-hoc explainability methods aim to provide explanations of what an already trained model, usually a neural network, learned and how it uses this information to arrive at a prediction. While not making a black box model fully transparent, by providing a glimpse into the black box these methods have the great advantage that they do not compromise performance, which is a desiderata for a neural network in practice. A majority of post-hoc explanation approaches, however, focus on instance-specific explanations, giving insights into why a decision was made for a particular instance (e.g., saliency maps, GradCam, Integrated Gradients, LIME, …). We are interested in global explanations that describe the general information encoded for example by a group of neurons and discovering such feature-encoding neuron groups in the first place and provide these along with theoretical guarantees.
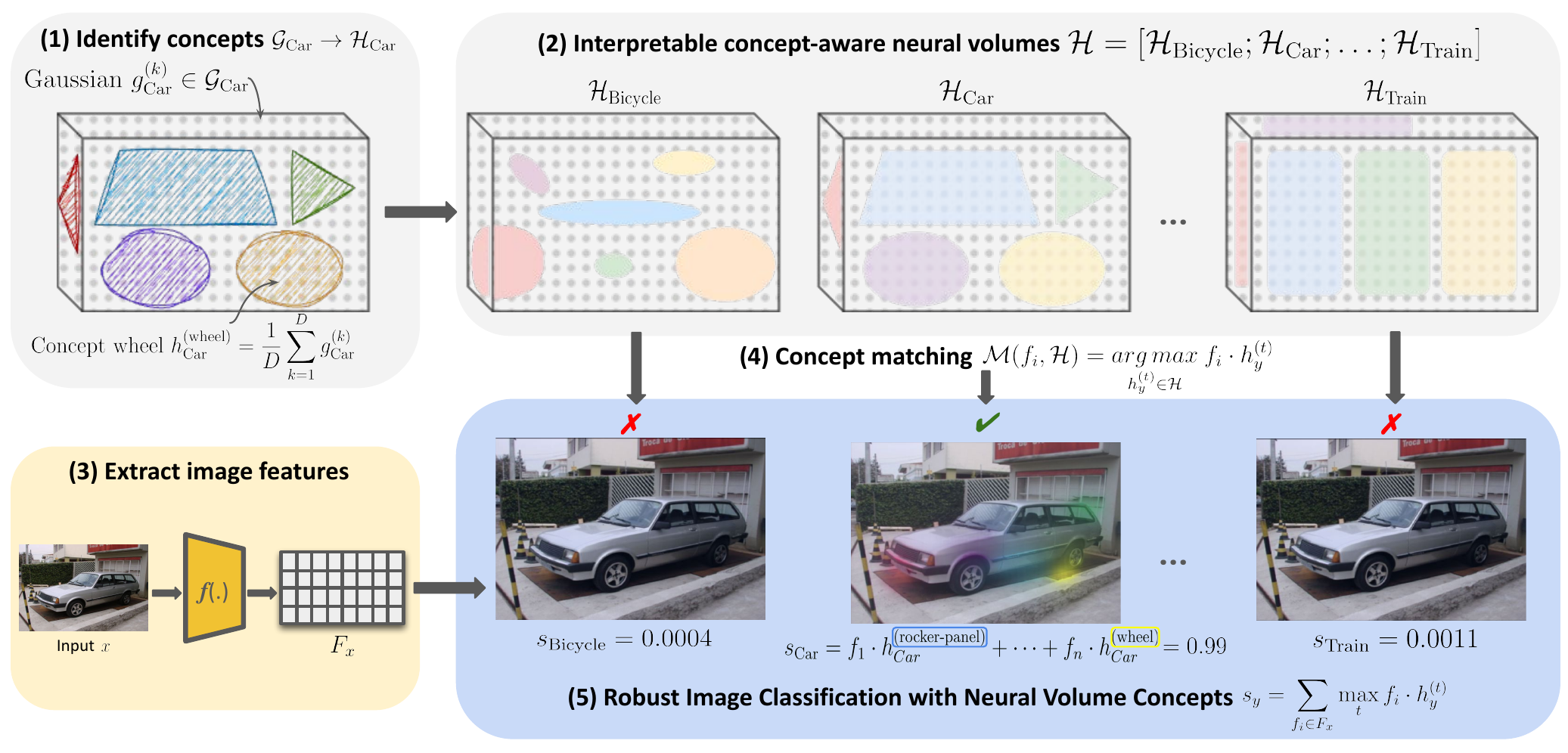
Inherently Interpretable decision-making
Inherently interpretable models are key to ensure explanations that are faithful to the actual model decisions. Such Explanations that reveal the true decision-making of the model are key in high-stakes settings in practice, for example in biomedical imaging – e.g., predicting cancer risk or events based on radiological images, tissue stains, or skin screens. We are interested in developing new methodology that yields both highly performant but still inherently interpretable models, ensure that explanations are correctly grounded in the input image, as well as models that are inherently interpretabl while emitting other properties such as OOD robustness.

Neuro-symbolic architectures
Neuro-symbolic approaches to machine learning promise full interpretability through symbolic encodings in a neural network that can be parsed and understood by a human domain expert. While highly interesting, these approaches remain largely theoretical, as such architectures are hard to define and even harder to optimize. We focus on recent advances for binarized and ternarized neural network architectures from different fields, such as ML for embedded devices, and how we can transfer these to neuro-symbolic learning. In several biological applications as well as settings with transactional data, such architectures already provide a fully interpretable approach that allows to derive new insights directly from the network.

Sparse Neural Networks
Sparse neural networks not only provide more resource-efficient alternatives to their dense counterparts at same performance, captured by the Lottery Ticket Hypothesis, but also represent useful information in their network topology compared to the standard fully connected layers in dense networks. We focus on the question of how we can prune networks to extreme sparsity, and how such sparse neural networks can then yield more explainable decision-making.
List of Publications
2025
Hedderich, MA, Wang, A, Zhao, R, Eichin, F, Fischer, J, Plank, B, What’s the Difference? Supporting Users in Identifying the Effects of Prompt and Model Changes Through Token Patterns ACL, 2025. [preprint]
Pham, N, Schiele, B, Kortylewski, A*, Fischer, J*, Escaping Plato’s Cave: Robust Conceptual Reasoning through Interpretable 3D Neural Object Volumes preprint: arXiv:2503.13429, 2025. [preprint] [project page] *equal contribution
Walter, NP, Vreeken, J, Fischer, J, Now you see me! A framework for obtaining class-relevant saliency maps preprint: arXiv:2503.07346, 2025. [preprint] [project page]
Görgün, A, Schiele, B, Fischer, J, VITAL: More Understandable Feature Visualization through Distribution Alignment and Relevant Information Flow preprint: arXiv:2503.22399, 2025. [preprint] [project page]
Sammani, F, Fischer, J, Deligiannis, N, Unlocking Open-Set Language Accessibility in Vision Models preprint: arXiv:2503.10981, 2025. [preprint]
Chen, C, Saha, E, Fischer, J, Guebila, MB, Fanfani, V, Shutta, K, Padi, M, Glass, K, DeMeo, D, Lopes-Ramos, C, Quackenbush, J, Identifying Sex Differences in Lung Adenocarcinoma Using Multi-Omics Integrative Protein Signaling Networks preprint: bioRxiv:2025.02.03.636354 [preprint]
Hesse, R, Fischer, J, Schaub-Meyer, S, Roth, S, Disentangling Polysemantic Channels in Convolutional Neural Networks accepted at CVPR workshop on Mechanistic Interpretability for Vision (MIV) 2025.
2024
Hossain, I‡, Fischer, J‡, Burkholz, R*, Quackenbush, J*, Pruning neural network models for gene regulatory dynamics using data and domain knowledge accepted at NeurIPS 2024. (25.8% acceptance rate, Core A*) [preprint]
‡*equal contribution
Fischer, J, Ma, R, Sailing in high-dimensional spaces: Low-dimensional embeddings through angle preservation preprint: arXiv:2406.09876, 2024. [preprint]
Fanfani, V, Shutta, KH, Mandros, P, Fischer, J, Saha, E, Micheletti, S, Chen, C, Guebila, MB, Lopes-Ramos, CM, Quackenbush, J, Reproducible processing of TCGA regulatory networks preprint: bioRxiv:2024.11.05.622163 [preprint]
Fischer, J, Shutta, KH, Chen, C, Fanfani, V, Saha, E, Mandros, P, Guebila, MB, Xiu, J, Nieva, J, Liu, S, Uprety, D, Spetzler, D, Lopes-Ramos, CM, DeMeo, D, Quackenbush, J, Selective loss of Y chromosomes in lung adenocarcinoma modulates the tumor immune environment through cancer/testis antigens bioRxiv:10.1101/2024.09.19.613876v1, 2024. [preprint]
Lin, Y, Breuer, K, Weichenhan, D, Lafrenz, P, Wilk, A, Chepeleva, M, Mücke, O, Schönung, M, Petermann, F, Kensche, P, Weiser, L, Thommen, F, Giacomelli, G, Nordstroem, K, Gonzales-Avalos, E, Merkel, A, Kretzmer, H, Fischer, J, Krämer, S, Iskar, M, Wolf, S, Buchhalter, I, Esteller, M, Lawerenz, C, Twardziok, S, Zapatka, M, Hovestadt, V, Schlesner, M, Schulz, M, Hoffmann, S, Gerhauser, C, Walter, J, Hartmann, M, Lipka, DB, Assenov, Y, Bock, C, Plass, C, Toth, R, Lutsik, P Pipeline Olympics: continuable benchmarking of computational workflows for DNA methylation sequencing data against an experimental gold-standard bioRxiv:10.1101/2024.09.16.609142, 2024. [preprint]
Mandros, P, Gallagher, I, Fanfani, V, Chen, C, Fischer, J, Ismail, A, Hsu, L, Saha, E, DeConti, DK, Quackenbush, J, node2vec2rank: Large Scale and Stable Graph Differential Analysis via Multi-Layer Node Embeddings and Ranking preprint: bioRxiv:10.1101/2024.06.16.599201v1, 2024. [preprint]
Walter, NP, Fischer, J, Vreeken, J, Finding Interpretable Class-Specific Patterns through Efficient Neural Search AAAI Conference on Artificial Intelligence (AAAI), 2024. (23.8% acceptance rate, Core A*) [Article]
Saha, E‡, Fanfani, V‡, Mandros, P, Guebila, MB, Fischer, J, Shutta, KH, Glass, K, DeMeo, DL, Lopes-Ramos, CM, Quackenbush, J, Bayesian inference of sample-specific coexpression networks Genome Research, CSHL, 2024. (IF: 6.70, 2022) [PDF]
‡equal contribution
Saha, E‡, Fanfani, V‡, Mandros, P, Guebila, MB, Fischer, J, Shutta, KH, Glass, K, DeMeo, DL, Lopes-Ramos, CM, Quackenbush, J, Bayesian Optimized sample-specific Networks Obtained By Omics data (BONOBO) Conference for Research in Computational Molecular Biology (RECOMB), 2024. (16.5% acceptance rate) [Conference paper]
‡equal contribution
Saha, E, Guebila, MB, Fanfani, V, Fischer, J, Shutta, KH, Mandros, P, DeMeo, DL, Quackenbush, J, Lopes-Ramos, CM, Gene regulatory Networks Reveal Sex Difference in Lung Adenocarcinoma Biology of Sex Differences 15(62), 2024. (IF: 8.24, 2022) [preprint] [Article]
Hossain, I, Fanfani, V, Fischer, J, Quackenbush, J, Burkholz, J, Biologically informed NeuralODEs for genome-wide regulatory dynamics Genome Biology 25(127), BMC, 2024. (IF: 17.4, 2022) [Article]
2023
Hedderich, M‡, Fischer, J‡, Klakow, D, Vreeken, J, Understanding and Mitigating Classification Errors Through Interpretable Token Patterns Empirical Methods in Natural Language Processing (EMNLP) BlackboxNLP workshop, 2023. [preprint]
‡equal contribution
Kamp, M, Fischer, J, Vreeken, J, Federated Learning from Small Datasets. International Conference on Learning Representations (ICLR), OpenReview, 2023. (31.8% acceptance rate, Core A*) [PDF]
Fischer, J, Burkholz, R, Vreeken, J, Preserving local densities in low-dimensional embeddings. preprint: arXiv:2301.13732, 2023. [preprint]
Fischer, J, Schulz, MH, Efficiently Quantifying DNA Methylation for Bulk- and Single-cell Bisulfite Data. Bioinformatics 39(6), Oxford University Press, 2023. (IF: 5.8, 2023) [Article]
2022
Hedderich, M‡, Fischer, J‡, Klakow, D, Vreeken, J, Label-Descriptive Patterns and their Application to Characterizing Classification Errors. In: Proceedings of the International Conference on Machine Learning (ICML), PMLR, 2022. (21.9% acceptance rate, Core A*) [PDF]
‡equal contribution
Fischer, J, Burkholz, R, Plant ‘n’ Seek: Can You Find the Winning Ticket? International Conference on Learning Representations (ICLR), OpenReview, 2022. (32.9% acceptance rate, Core A*) [PDF]
Marx, A, Fischer, J, Estimating Mutual Information via Geodesic kNN. SIAM Conference on Data Mining (SDM), SIAM, 2022. (27.9% acceptance rate, Core A) [Article]
2021
Fischer, J, Vreeken, J, Differentiable Pattern Set Mining. In: Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD), ACM, 2021. (15.4% acceptance rate, Core A*) [Article]
Fischer, J, Oláh, A, Vreeken, J, What’s in the Box? Explaining Neural Networks with Robust Rules. In: Proceedings of the International Conference on Machine Learning (ICML), PMLR, 2021. (21.4% acceptance rate, Core A*) [PDF]
Fischer, J, Ardakani, FB, Kattler, K, Walter, J, Schulz, MH, CpG content-dependent associations between transcription factors and histone modifications. Plos ONE 16(4): e0249985, 2021. (IF: 3.7, 2023) [Article]
Fischer, J‡, Gadhikar‡, A, Burkholz, R, Lottery Tickets with Nonzero Biases. preprint: arXiv:2110.11150, 2021. [preprint]
‡equal contribution
Heiter, E, Fischer, J, Vreeken, J, Factoring out prior knowledge from low-dimensional embeddings. preprint: arXiv:2103.01828, 2021. [preprint]
2020
Fischer, J, Vreeken, J, Discovering Succinct Pattern Sets Expressing Co-Occurrence and Mutual Exclusivity. In: Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD), ACM, 2020. (16.8% acceptance rate, Core A*) [Article]
2019
Fischer, J, Vreeken, J, Sets of Robust Rules, and How to Find Them In: Proceedings of the European Conference on Machine Learning and Principles and Practice of Knowledge Discovery in Data (ECMLPKDD), Springer, 2019. (17.7% acceptance rate, Core A, ==selected plenary talk==, <5% of accepted papers) [Article]
Fischer, J, Schulz, MH, Fast and accurate bisulfite alignment and methylation calling for mammalian genomes. Talk at ISMB/ECCB, Basel, Switzerland, 2019.
2018
Ardakani, FB Kattler, K, Nordström, KJ, Gasparoni, N, Gasparoni, G, Fuchs, S, Sinha, A, Barann, M, Ebert, P, Fischer, J, Hutter, B, Zipprich, G, Imbusch, CD, Felder, B, Eils, J, Brors, B, Lengauer, T, Manke, T, Rosenstiel, P, Walter, J, Schulz, MH, Integrative analysis of single-cell expression data reveals distinct regulatory states in bidirectional promoters. Epigenetics & Chromatin 11(1): 66, 2018. (IF: 5.5, 2023) [Article]
Fischer, J, Schulz, MH, Fast and accurate bisulfite alignment and methylation calling for mammalian genomes. Short talk/poster at RECOMB-seq/RECOMB, Paris, France, 2018.
Theses
Fischer, J, More than the sum of its parts – pattern mining, neural networks, and how they complement each other. Doctoral Dissertation, 2022. [PDF]